# Time series
# Plotting
import holoviews as hv
import panel as pn
import hvplot
import hvplot.pandas # noqa
# Apply extensions
hvplot.extension('bokeh')
# Analysis
import pandas as pd
import skchange # noqa
# Utility
import watermark
from skchange.datasets import generate_changing_data
The guide introduces the concepts of time series change detection and demonstrates practical applications using the Python programming language, primarily with the sktime and skchange libraries. Change detection identifies moments when a time series’ statistical properties shift; this underpins segmentation, point anomaly detection, and segment anomaly detection. sktime is described as a general-purpose time series toolkit with a scikit-learn-like Application Programing Interface (API), while skchange focuses on change-point detection and segmentation using statistical tests, kernel methods, and their efficient implementations (often via numba).
Using synthetic data with known changes, the guide shows how defaults can misalign detections and how adjusting detection parameters can improve results. It emphasizes inspecting underlying scores and per-sample labels to guide calibration of these parameters rather than blind trial-and-error.
skchange and sktime
sktime [1] is a flexible tool for time series analysis, providing a comprehensive toolkit that mirrors scikit-learn’s approach but for temporal data. It handles the full spectrum of time series tasks including forecasting with methods ranging from classical ARIMA [2] to modern deep learning models, time series classification and regression, clustering, and essential preprocessing transformations. It maintains the same unified API design as sklearn and therefore makes it easy to experiment with different algorithms and build robust time series machine learning pipelines.
skchange [3] is more focused, specializing in exclusively Change Point detection and Time Series Segmentation. This library attempts to identify the moment when statistical properties shift in the data — detecting changes in mean, variance, or the underlying distribution using various statistical tests, kernel methods, and machine learning techniques. skchange is therefore useful in applications like monitoring system performance degradation, spotting regime changes in financial markets, or identifying behavioral shifts in Internet-of-Things (IoT) sensor streams.
The documentation for skchange is currently in development as well as its integration into sktime and therefore one of the motivating factors for this post was to make it more widely known until such time the documentation matures. It is based on a series of tutorials provided by the authors most notably the PyData Global 2024 Workshop Notebook Tutorials [4]. This has been superseded by the skchange tutorial at Hydro Data Science Forum.
Environment Setup
Create a conda environment using the mamba [5] tool (or a virtual environment manager of your choice) and install a Python 3.12 environment. Install the following initial packages:
pipsktimeseabornpmdarimastatsmodelsnumba
# Create a new environment
mamba create -n sktime python=3.12
Looking for: ['python=3.12']
conda-forge/linux-64 Using cache
conda-forge/noarch Using cache
Transaction
Prefix: /home/miah0x41/mambaforge/envs/sktime
Updating specs:
- python=3.12
Package Version Build Channel Size
─────────────────────────────────────────────────────────────────────────────
Install:
─────────────────────────────────────────────────────────────────────────────
+ ld_impl_linux-64 2.43 h712a8e2_4 conda-forge Cached
+ _libgcc_mutex 0.1 conda_forge conda-forge Cached
+ libgomp 15.1.0 h767d61c_2 conda-forge Cached
+ _openmp_mutex 4.5 2_gnu conda-forge Cached
+ libgcc 15.1.0 h767d61c_2 conda-forge Cached
+ ncurses 6.5 h2d0b736_3 conda-forge Cached
+ libzlib 1.3.1 hb9d3cd8_2 conda-forge Cached
+ liblzma 5.8.1 hb9d3cd8_1 conda-forge Cached
+ libgcc-ng 15.1.0 h69a702a_2 conda-forge Cached
+ libffi 3.4.6 h2dba641_1 conda-forge Cached
+ libexpat 2.7.0 h5888daf_0 conda-forge Cached
+ readline 8.2 h8c095d6_2 conda-forge Cached
+ libsqlite 3.49.2 hee588c1_0 conda-forge Cached
+ tk 8.6.13 noxft_h4845f30_101 conda-forge Cached
+ libxcrypt 4.4.36 hd590300_1 conda-forge Cached
+ bzip2 1.0.8 h4bc722e_7 conda-forge Cached
+ libuuid 2.38.1 h0b41bf4_0 conda-forge Cached
+ libnsl 2.0.1 hd590300_0 conda-forge Cached
+ tzdata 2025b h78e105d_0 conda-forge Cached
+ ca-certificates 2025.4.26 hbd8a1cb_0 conda-forge Cached
+ openssl 3.5.0 h7b32b05_1 conda-forge Cached
+ python 3.12.10 h9e4cc4f_0_cpython conda-forge Cached
+ wheel 0.45.1 pyhd8ed1ab_1 conda-forge Cached
+ setuptools 80.8.0 pyhff2d567_0 conda-forge Cached
+ pip 25.1.1 pyh8b19718_0 conda-forge Cached
Summary:
Install: 25 packages
Total download: 0 B
─────────────────────────────────────────────────────────────────────────────
Confirm changes: [Y/n]
Downloading and Extracting Packages:
Preparing transaction: done
Verifying transaction: done
Executing transaction: done
To activate this environment, use
$ mamba activate sktime
To deactivate an active environment, use
$ mamba deactivate
# Activate environment
mamba activate sktime
# Install sktime
mamba install -c conda-forge pip sktime seaborn pmdarima statsmodels numba
Looking for: ['pip', 'sktime', 'seaborn', 'pmdarima', 'statsmodels', 'numba']
conda-forge/linux-64 Using cache
conda-forge/noarch Using cache
Pinned packages:
- python 3.12.*
Transaction
Prefix: /home/miah0x41/mambaforge/envs/sktime
Updating specs:
- pip
- sktime
- seaborn
- pmdarima
- statsmodels
- numba
- ca-certificates
- openssl
Package Version Build Channel Size
────────────────────────────────────────────────────────────────────────────────
Install:
────────────────────────────────────────────────────────────────────────────────
+ libstdcxx 15.1.0 h8f9b012_2 conda-forge 4MB
+ libpng 1.6.47 h943b412_0 conda-forge 289kB
+ libgfortran5 15.1.0 hcea5267_2 conda-forge 2MB
+ libjpeg-turbo 3.1.0 hb9d3cd8_0 conda-forge 629kB
+ libwebp-base 1.5.0 h851e524_0 conda-forge 430kB
+ pthread-stubs 0.4 hb9d3cd8_1002 conda-forge 8kB
+ xorg-libxdmcp 1.1.5 hb9d3cd8_0 conda-forge 20kB
+ xorg-libxau 1.0.12 hb9d3cd8_0 conda-forge 15kB
+ libdeflate 1.24 h86f0d12_0 conda-forge 73kB
+ libbrotlicommon 1.1.0 hb9d3cd8_2 conda-forge Cached
+ zstd 1.5.7 hb8e6e7a_2 conda-forge 568kB
+ lerc 4.0.0 h0aef613_1 conda-forge 264kB
+ libstdcxx-ng 15.1.0 h4852527_2 conda-forge 35kB
+ libfreetype6 2.13.3 h48d6fc4_1 conda-forge 380kB
+ libgfortran 15.1.0 h69a702a_2 conda-forge 35kB
+ libxcb 1.17.0 h8a09558_0 conda-forge 396kB
+ libbrotlienc 1.1.0 hb9d3cd8_2 conda-forge Cached
+ libbrotlidec 1.1.0 hb9d3cd8_2 conda-forge Cached
+ libtiff 4.7.0 hf01ce69_5 conda-forge 430kB
+ qhull 2020.2 h434a139_5 conda-forge 553kB
+ libfreetype 2.13.3 ha770c72_1 conda-forge 8kB
+ libopenblas 0.3.29 pthreads_h94d23a6_0 conda-forge Cached
+ brotli-bin 1.1.0 hb9d3cd8_2 conda-forge 19kB
+ openjpeg 2.5.3 h5fbd93e_0 conda-forge 343kB
+ lcms2 2.17 h717163a_0 conda-forge 248kB
+ freetype 2.13.3 ha770c72_1 conda-forge 172kB
+ libblas 3.9.0 31_h59b9bed_openblas conda-forge Cached
+ brotli 1.1.0 hb9d3cd8_2 conda-forge 19kB
+ libcblas 3.9.0 31_he106b2a_openblas conda-forge Cached
+ liblapack 3.9.0 31_h7ac8fdf_openblas conda-forge Cached
+ python_abi 3.12 7_cp312 conda-forge 7kB
+ packaging 25.0 pyh29332c3_1 conda-forge 62kB
+ scikit-base 0.12.2 pyhecae5ae_0 conda-forge 110kB
+ joblib 1.4.2 pyhd8ed1ab_1 conda-forge Cached
+ threadpoolctl 3.6.0 pyhecae5ae_0 conda-forge 24kB
+ pytz 2025.2 pyhd8ed1ab_0 conda-forge 189kB
+ python-tzdata 2025.2 pyhd8ed1ab_0 conda-forge 144kB
+ cycler 0.12.1 pyhd8ed1ab_1 conda-forge 13kB
+ pyparsing 3.2.3 pyhd8ed1ab_1 conda-forge 96kB
+ munkres 1.1.4 pyh9f0ad1d_0 conda-forge Cached
+ hpack 4.1.0 pyhd8ed1ab_0 conda-forge Cached
+ hyperframe 6.1.0 pyhd8ed1ab_0 conda-forge Cached
+ pysocks 1.7.1 pyha55dd90_7 conda-forge Cached
+ six 1.17.0 pyhd8ed1ab_0 conda-forge Cached
+ pycparser 2.22 pyh29332c3_1 conda-forge Cached
+ h2 4.2.0 pyhd8ed1ab_0 conda-forge Cached
+ python-dateutil 2.9.0.post0 pyhff2d567_1 conda-forge Cached
+ unicodedata2 16.0.0 py312h66e93f0_0 conda-forge 404kB
+ brotli-python 1.1.0 py312h2ec8cdc_2 conda-forge Cached
+ pillow 11.2.1 py312h80c1187_0 conda-forge 43MB
+ kiwisolver 1.4.8 py312h84d6215_0 conda-forge 72kB
+ llvmlite 0.44.0 py312h374181b_1 conda-forge 30MB
+ cython 3.1.1 py312h2614dfc_1 conda-forge 4MB
+ numpy 2.2.6 py312h72c5963_0 conda-forge 8MB
+ cffi 1.17.1 py312h06ac9bb_0 conda-forge Cached
+ fonttools 4.58.0 py312h178313f_0 conda-forge 3MB
+ contourpy 1.3.2 py312h68727a3_0 conda-forge 277kB
+ scipy 1.15.2 py312ha707e6e_0 conda-forge 17MB
+ pandas 2.2.3 py312hf9745cd_3 conda-forge 15MB
+ numba 0.61.2 py312h2e6246c_0 conda-forge 6MB
+ zstandard 0.23.0 py312h66e93f0_2 conda-forge 732kB
+ matplotlib-base 3.10.3 py312hd3ec401_0 conda-forge 8MB
+ scikit-learn 1.6.1 py312h7a48858_0 conda-forge 11MB
+ sktime 0.36.0 py312h7900ff3_0 conda-forge 34MB
+ patsy 1.0.1 pyhd8ed1ab_1 conda-forge 187kB
+ urllib3 2.4.0 pyhd8ed1ab_0 conda-forge 101kB
+ seaborn-base 0.13.2 pyhd8ed1ab_3 conda-forge 228kB
+ statsmodels 0.14.4 py312hc0a28a1_0 conda-forge 12MB
+ pmdarima 2.0.4 py312h41a817b_2 conda-forge 663kB
+ seaborn 0.13.2 hd8ed1ab_3 conda-forge 7kB
Summary:
Install: 70 packages
Total download: 205MB
────────────────────────────────────────────────────────────────────────────────
Confirm changes: [Y/n]
libpng 288.7kB @ 552.8kB/s 0.5s
libgfortran5 1.6MB @ 3.0MB/s 0.5s
libjpeg-turbo 628.9kB @ 1.2MB/s 0.5s
lerc 264.2kB @ 450.8kB/s 0.1s
libstdcxx-ng 34.6kB @ 57.8kB/s 0.1s
libfreetype6 380.1kB @ 626.1kB/s 0.1s
libstdcxx 3.9MB @ 6.3MB/s 0.6s
libwebp-base 430.0kB @ 689.0kB/s 0.6s
openjpeg 343.0kB @ 538.3kB/s 0.1s
lcms2 248.0kB @ 346.6kB/s 0.1s
freetype 172.4kB @ 240.1kB/s 0.1s
threadpoolctl 23.9kB @ 33.2kB/s 0.1s
pytz 189.0kB @ 261.6kB/s 0.1s
contourpy 276.5kB @ 268.9kB/s 0.3s
cython 3.7MB @ 2.9MB/s 0.6s
fonttools 2.8MB @ 2.1MB/s 0.6s
pthread-stubs 8.3kB @ 6.0kB/s 0.1s
libdeflate 72.6kB @ 51.7kB/s 0.1s
libxcb 395.9kB @ 270.8kB/s 0.1s
libtiff 429.6kB @ 280.9kB/s 0.1s
python_abi 7.0kB @ 4.5kB/s 0.1s
python-tzdata 144.2kB @ 89.0kB/s 0.1s
unicodedata2 404.4kB @ 243.3kB/s 0.1s
numpy 8.5MB @ 4.9MB/s 1.0s
zstandard 732.2kB @ 406.8kB/s 0.1s
seaborn-base 227.8kB @ 121.4kB/s 0.1s
xorg-libxdmcp 19.9kB @ 10.1kB/s 0.1s
libgfortran 34.5kB @ 16.5kB/s 0.1s
brotli-bin 18.9kB @ 8.6kB/s 0.1s
scikit-base 110.2kB @ 45.8kB/s 0.2s
scipy 17.1MB @ 4.9MB/s 1.9s
llvmlite 30.0MB @ 8.6MB/s 2.8s
patsy 186.6kB @ 52.9kB/s 0.0s
seaborn 6.9kB @ 1.9kB/s 0.0s
qhull 552.9kB @ 151.8kB/s 0.1s
packaging 62.5kB @ 16.9kB/s 0.0s
kiwisolver 71.6kB @ 18.9kB/s 0.1s
numba 5.9MB @ 1.5MB/s 0.5s
xorg-libxau 14.8kB @ 3.7kB/s 0.1s
statsmodels 12.1MB @ 3.0MB/s 3.0s
brotli 19.3kB @ 4.7kB/s 0.1s
pmdarima 663.1kB @ 158.9kB/s 0.1s
cycler 13.4kB @ 3.2kB/s 0.1s
urllib3 100.8kB @ 23.3kB/s 0.1s
scikit-learn 10.6MB @ 2.4MB/s 0.6s
pyparsing 96.0kB @ 21.9kB/s 0.0s
libfreetype 7.7kB @ 1.7kB/s 0.0s
zstd 567.6kB @ 126.1kB/s 0.1s
matplotlib-base 8.2MB @ 1.6MB/s 0.6s
pillow 42.5MB @ 8.5MB/s 2.6s
pandas 15.4MB @ 2.8MB/s 1.5s
sktime 34.1MB @ 3.8MB/s 7.5s
Downloading and Extracting Packages:
Preparing transaction: done
Verifying transaction: done
Executing transaction: done
# Install Skchange
pip install skchange[numba]
Collecting skchange[numba]
Downloading skchange-0.13.0-py3-none-any.whl.metadata (5.6 kB)
Requirement already satisfied: numpy>=1.21 in /home/miah0x41/mambaforge/envs/sktime/lib/python3.12/site-packages (from skchange[numba]) (2.2.6)
Requirement already satisfied: pandas>=1.1 in /home/miah0x41/mambaforge/envs/sktime/lib/python3.12/site-packages (from skchange[numba]) (2.2.3)
Requirement already satisfied: sktime>=0.35 in /home/miah0x41/mambaforge/envs/sktime/lib/python3.12/site-packages (from skchange[numba]) (0.36.0)
Requirement already satisfied: numba<0.62,>=0.61 in /home/miah0x41/mambaforge/envs/sktime/lib/python3.12/site-packages (from skchange[numba]) (0.61.2)
Requirement already satisfied: llvmlite<0.45,>=0.44.0dev0 in /home/miah0x41/mambaforge/envs/sktime/lib/python3.12/site-packages (from numba<0.62,>=0.61->skchange[numba]) (0.44.0)
Requirement already satisfied: python-dateutil>=2.8.2 in /home/miah0x41/mambaforge/envs/sktime/lib/python3.12/site-packages (from pandas>=1.1->skchange[numba]) (2.9.0.post0)
Requirement already satisfied: pytz>=2020.1 in /home/miah0x41/mambaforge/envs/sktime/lib/python3.12/site-packages (from pandas>=1.1->skchange[numba]) (2025.2)
Requirement already satisfied: tzdata>=2022.7 in /home/miah0x41/mambaforge/envs/sktime/lib/python3.12/site-packages (from pandas>=1.1->skchange[numba]) (2025.2)
Requirement already satisfied: six>=1.5 in /home/miah0x41/mambaforge/envs/sktime/lib/python3.12/site-packages (from python-dateutil>=2.8.2->pandas>=1.1->skchange[numba]) (1.17.0)
Requirement already satisfied: joblib<1.5,>=1.2.0 in /home/miah0x41/mambaforge/envs/sktime/lib/python3.12/site-packages (from sktime>=0.35->skchange[numba]) (1.4.2)
Requirement already satisfied: packaging in /home/miah0x41/mambaforge/envs/sktime/lib/python3.12/site-packages (from sktime>=0.35->skchange[numba]) (25.0)
Requirement already satisfied: scikit-base<0.13.0,>=0.6.1 in /home/miah0x41/mambaforge/envs/sktime/lib/python3.12/site-packages (from sktime>=0.35->skchange[numba]) (0.12.2)
Requirement already satisfied: scikit-learn<1.7.0,>=0.24 in /home/miah0x41/mambaforge/envs/sktime/lib/python3.12/site-packages (from sktime>=0.35->skchange[numba]) (1.6.1)
Requirement already satisfied: scipy<2.0.0,>=1.2 in /home/miah0x41/mambaforge/envs/sktime/lib/python3.12/site-packages (from sktime>=0.35->skchange[numba]) (1.15.2)
Requirement already satisfied: threadpoolctl>=3.1.0 in /home/miah0x41/mambaforge/envs/sktime/lib/python3.12/site-packages (from scikit-learn<1.7.0,>=0.24->sktime>=0.35->skchange[numba]) (3.6.0)
Downloading skchange-0.13.0-py3-none-any.whl (18.4 MB)
━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━ 18.4/18.4 MB 8.6 MB/s eta 0:00:00
Installing collected packages: skchange
Successfully installed skchange-0.13.0Install the skchange package with pip and the following additional packages with conda or mamba:
# Install Skchange
pip install skchange[numba]
Collecting skchange[numba]
Downloading skchange-0.13.0-py3-none-any.whl.metadata (5.6 kB)
Requirement already satisfied: numpy>=1.21 in /home/miah0x41/mambaforge/envs/sktime/lib/python3.12/site-packages (from skchange[numba]) (2.2.6)
Requirement already satisfied: pandas>=1.1 in /home/miah0x41/mambaforge/envs/sktime/lib/python3.12/site-packages (from skchange[numba]) (2.2.3)
Requirement already satisfied: sktime>=0.35 in /home/miah0x41/mambaforge/envs/sktime/lib/python3.12/site-packages (from skchange[numba]) (0.36.0)
Requirement already satisfied: numba<0.62,>=0.61 in /home/miah0x41/mambaforge/envs/sktime/lib/python3.12/site-packages (from skchange[numba]) (0.61.2)
Requirement already satisfied: llvmlite<0.45,>=0.44.0dev0 in /home/miah0x41/mambaforge/envs/sktime/lib/python3.12/site-packages (from numba<0.62,>=0.61->skchange[numba]) (0.44.0)
Requirement already satisfied: python-dateutil>=2.8.2 in /home/miah0x41/mambaforge/envs/sktime/lib/python3.12/site-packages (from pandas>=1.1->skchange[numba]) (2.9.0.post0)
Requirement already satisfied: pytz>=2020.1 in /home/miah0x41/mambaforge/envs/sktime/lib/python3.12/site-packages (from pandas>=1.1->skchange[numba]) (2025.2)
Requirement already satisfied: tzdata>=2022.7 in /home/miah0x41/mambaforge/envs/sktime/lib/python3.12/site-packages (from pandas>=1.1->skchange[numba]) (2025.2)
Requirement already satisfied: six>=1.5 in /home/miah0x41/mambaforge/envs/sktime/lib/python3.12/site-packages (from python-dateutil>=2.8.2->pandas>=1.1->skchange[numba]) (1.17.0)
Requirement already satisfied: joblib<1.5,>=1.2.0 in /home/miah0x41/mambaforge/envs/sktime/lib/python3.12/site-packages (from sktime>=0.35->skchange[numba]) (1.4.2)
Requirement already satisfied: packaging in /home/miah0x41/mambaforge/envs/sktime/lib/python3.12/site-packages (from sktime>=0.35->skchange[numba]) (25.0)
Requirement already satisfied: scikit-base<0.13.0,>=0.6.1 in /home/miah0x41/mambaforge/envs/sktime/lib/python3.12/site-packages (from sktime>=0.35->skchange[numba]) (0.12.2)
Requirement already satisfied: scikit-learn<1.7.0,>=0.24 in /home/miah0x41/mambaforge/envs/sktime/lib/python3.12/site-packages (from sktime>=0.35->skchange[numba]) (1.6.1)
Requirement already satisfied: scipy<2.0.0,>=1.2 in /home/miah0x41/mambaforge/envs/sktime/lib/python3.12/site-packages (from sktime>=0.35->skchange[numba]) (1.15.2)
Requirement already satisfied: threadpoolctl>=3.1.0 in /home/miah0x41/mambaforge/envs/sktime/lib/python3.12/site-packages (from scikit-learn<1.7.0,>=0.24->sktime>=0.35->skchange[numba]) (3.6.0)
Downloading skchange-0.13.0-py3-none-any.whl (18.4 MB)
━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━ 18.4/18.4 MB 8.6 MB/s eta 0:00:00
Installing collected packages: skchange
Successfully installed skchange-0.13.0Additional packages are from the skchange tutorial based on the workshop at PyData Global 2024. An overview of the tutorial is available on YouTube:
The tutorial notebooks require the use of plotly and in particular the Plotly Express package as well as the nbformat package.
For the correct display of Plotly plots where jupyter lab is provisioned by a different Python environment, plotly must also be installed in this environment as per GitHub Issue 4354 and reported in the Plotly forums [6].
The packages required for the tutorials (and built in plotting) are:
nbformatplotly
Additional recommend packages are:
If using a Jupyter environment then the ipykernel package is recommended; register the new environment (and in this case called sktime) with the following:
# Register kernel
python -m ipykernel install --user --name sktime --display-name "sktime (py3.12)"
Installed kernelspec sktime in /home/miah0x41/.local/share/jupyter/kernels/sktimeSystem Details
Import packages:
# System version
print(watermark.watermark())Last updated: 2025-08-31T16:16:55.187357+01:00
Python implementation: CPython
Python version : 3.12.10
IPython version : 9.2.0
Compiler : GCC 13.3.0
OS : Linux
Release : 6.6.87.2-microsoft-standard-WSL2
Machine : x86_64
Processor : x86_64
CPU cores : 20
Architecture: 64bit
# Package versions
print(watermark.watermark(iversions=True, globals_=globals()))holoviews: 1.20.2
hvplot : 0.11.3
pandas : 2.2.3
panel : 1.7.0
skchange : 0.13.0
watermark: 2.5.0
Detection Systems
There are four types of detection tasks for time series:
- Change Detection - identify where a fundamental change has occurred marked by Change Points
- Segmentation - using Change Detection to segment the data with common attributes
- Point Anomaly Detection - using Change Detection to identify single points that are different
- Segment Anomaly Detection - using Change Detection to identify segments that are different
The foundation is Change Detection and the figure below illustrates the concepts of Change Points and the target of Change Detection systems:
This was generated using the helper function generate_changing_data from the skchange.datasets package and illustrates a single variable with two distinct segments. The red lines indicate where a change in the time series has occurred. The time period between these points reflects a Segment and in this instance reflects an Anamolous Segment.
The example time series uses randomised data around a mean of 0, -3 & 0 respectively and changes at time steps 50 and 150; it is entirely artificial and therefore fake data with known characteristics. Given it’s construction any function that discriminates using the mean should be able to identify the two types of segments.
Change Detection
The skchange package offers the following detectors:
MovingWindow-
“A generalized version of the MOSUM (moving sum) algorithm for changepoint detection. It runs a test statistic for a single changepoint at the midpoint in a moving window of length
2 * bandwidthover the data. Efficiently implemented using numba.” [7] PELT- “Pruned exact linear time changepoint detection, from skchange.” [8]
SeededBinarySegmentation- “Binary segmentation type changepoint detection algorithms recursively split the data into two segments, and test whether the two segments are different. The seeded binary segmentation algorithm is an efficient version of such algorithms, which tests for changepoints in intervals of exponentially growing length. It has the same theoretical guarantees as the original binary segmentation algorithm, but runs in log-linear time no matter the changepoint configuration.” [9]
The detector is in effect the mechanism an algorithm is applied across the time series data and usually consists of a parameter called bandwidth, which determines the extent of a window used to evaluate whether a change has occurred. The mechanism to determine if a change has occurred is typically done with a scoring or cost function - the idea that substantial changes in these metrics indicates a change in the underlying data and hence a Change Point.
We’ll start with the MovingWindow detector, which compares a number of data points (bandwidth) on either side of a split, using a scoring or cost mechanism (change_score) to compare the two sides and a detection threshold (penalty).
change_score: Anskchangeobject that measure the difference between two data intervals.bandwidth: The number of data points on either side of the split in the window.penalty: The detection threshold. The higher, the fewer detected change points. We will use the default throughout this tutorial.
The default change_score is L2cost, which is a term used in optimisation and Machine Learning for the Mean Squared Error (MSE) [10].
# Import detector
from skchange.change_detectors import MovingWindow
from skchange.costs import L2CostExpand the interactive sktime module interface to examine the detector to confirm the parameters in the code.
# Define detector (with defaults)
detector = MovingWindow(
change_score=L2Cost(),
bandwidth=30,
penalty=None,
)
# Preview
detectorMovingWindow(bandwidth=30, change_score=L2Cost())Please rerun this cell to show the HTML repr or trust the notebook.
MovingWindow(bandwidth=30, change_score=L2Cost())
L2Cost()
L2Cost()
# Fit the detector
detector = detector.fit(df)We can see below that the second detected change point is out by a single time step (i.e. 149 vs 150).
# Predict / apply detector
detections = detector.predict(df)
# Preview results
detections| ilocs | |
|---|---|
| 0 | 50 |
| 1 | 149 |
The figure below shows a small misalignment between the MovingWindow detector and the actual Change Point 2.
This illustrates the most common challenge with these tools and that’s the selection of the input or hyperparameters to refine the detector. The most likely candidate for refining the detector is the bandwidth to better capture the period of interest. In the following example, the bandwidth is halved to 15.
# Define detector
detector = MovingWindow(
change_score=L2Cost(),
bandwidth=15,
penalty=None,
)
# Fit the detector
detector = detector.fit(df)
# Predict / apply detector
detections = detector.predict(df)Changing the bandwidth from 30 to 15 steps appears to have improved the detection of the Change Points. In practice, we won’t know the true change points and therefore it is insufficent to apply the detector without some calibration activity.
Rather than using a Cost function we can also try a Change Score like CUSUM which measures the difference in the mean either side of the split:
# Import the CUSUM score
from skchange.change_scores import CUSUM# Define detector
detector = MovingWindow(
change_score=CUSUM(),
bandwidth=30,
penalty=None,
)
# Fit the detector
detector = detector.fit(df)
# Predict / apply detector
detections = detector.predict(df)Changing the change_score to CUSUM with the default bandwidth results with only a single change point being detected!
Let’s attempt the same trick and reduce the bandwidth from 30 to 15:
# Define detector
detector = MovingWindow(
change_score=CUSUM(),
bandwidth=15,
penalty=None,
)
# Fit the detector
detector = detector.fit(df)
# Predict / apply detector
detections = detector.predict(df)We appear to have made things worse as none of the points are detected!
The random selection of scoring/cost metrics and input parameters is not a robust approach to setting up a detector for even a trivial timeseries example such as this.
Change Scores Introspection
Whilst some users will advocate a “trial and error” approach or a systemic change in the input parameters to attempt to understand what changes are required to detect changes of interest, it is unreliable and time consuming. We can examine the underlying scores to make more informed choices on how to change the input parameters using a two step process:
- Plot the change point labels against time
- Plot the underlying score/cost metric
L2Cost
In this section we compare two MovingWindow detectors, which by default employ the L2Cost metric with the two bandwidth options of 30 and 15:
# Define detector for bw=30
d1 = MovingWindow(
change_score=L2Cost(),
bandwidth=30,
penalty=None,
)
# Fit the detector
d1 = d1.fit(df)
# Extract row-wise labels
bw30_labels = d1.transform(df)
# Preview
bw30_labels.loc[bw30_labels.labels > 0].sample(10, random_state=23)| labels | |
|---|---|
| 217 | 2 |
| 64 | 1 |
| 114 | 1 |
| 169 | 2 |
| 208 | 2 |
| 174 | 2 |
| 88 | 1 |
| 127 | 1 |
| 117 | 1 |
| 292 | 2 |
Repeat the process for the variation in the costs.
# Get scores/costs
bw30_scores = d1.transform_scores(df)
bw15_scores = d2.transform_scores(df)Once the plots have been generated using hvplot, convert to holoviews objects and combine their x-axes.
# Convert to bokeh and share x-axis
bokeh_p1 = hv.render(plot_labels)
bokeh_p2 = hv.render(plot_scores)
bokeh_p2.x_range = bokeh_p1.x_range # Share x-axis
# Stretch plots
bokeh_p1.sizing_mode = 'stretch_width'
bokeh_p2.sizing_mode = 'stretch_width'
# Create panel layout
dashboard = pn.Column(
pn.pane.Bokeh(bokeh_p1),
pn.Spacer(height=20),
pn.pane.Bokeh(bokeh_p2),
sizing_mode='stretch_width'
)The first plot shows the last active label for each time step. The series begins with label 0 and at the first change point increases to 1 and then followed by 2 after the second change point. This mechanism also highlights how the data could be segmented from this data set alone based on the label assigned, however it does not convery that labels 0 and 2 are the same.
The second plot shows the L2Cost for both detections; the red line for a the wider bandwidth shows a larger peak and wider based than the lower bandwidth of 15. The key difference is the position of the peak.
Change in Label and Cost
Zooming in shows that for the red line, it peaks at time step 149 and not at 150.
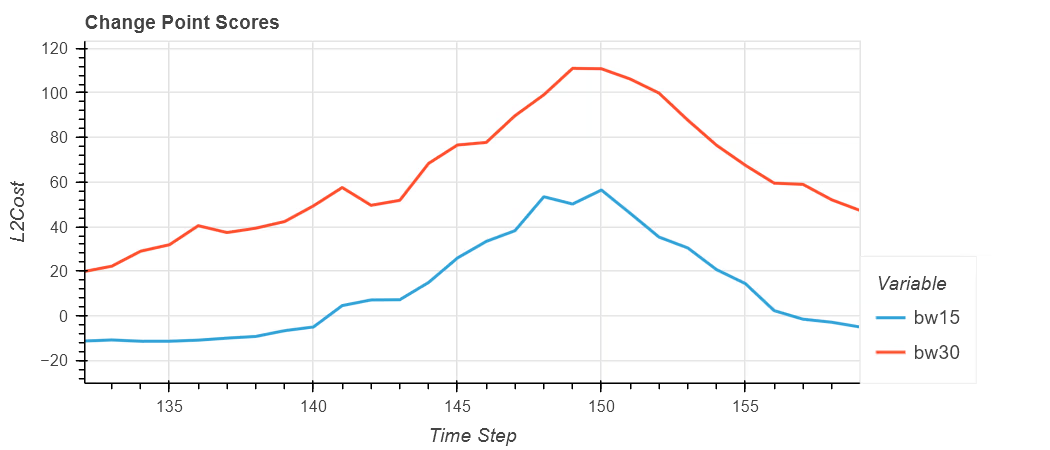
It’s important to note that there is no other evidence present to help indicate that the position of the change point is incorrect other than comparing the results to the original data and making a judgement to the suitability of the choices made.
CUSUM
A method from the 1950s that looks to compare the mean either side of a split window.
# Define detector for bw=30
d1 = MovingWindow(
change_score=CUSUM(),
bandwidth=30,
penalty=None,
)
# Fit the detector
d1 = d1.fit(df)
# Extract row-wise labels
bw30_labels = d1.transform(df)
# Preview
bw30_labels.loc[bw30_labels.labels > 0].sample(10, random_state=23)| labels | |
|---|---|
| 217 | 1 |
| 64 | 1 |
| 114 | 1 |
| 169 | 1 |
| 208 | 1 |
| 174 | 1 |
| 88 | 1 |
| 127 | 1 |
| 117 | 1 |
| 292 | 1 |
The first plot is we expect that only the CUSUM score based detector with a bandwidth of 30 was able to detect a change point. The second plot shows CUSUM values similar in shape to the L2Cost plots but note that only the red line at the first peak has a value greater than zero. The second peak and both peaks in blue do not suggesting that we require a change in a threshold or level to make the detector work.
It’s worth noting the similarities in shape between the two methods i.e. L2Cost and CUSUM. This indicates that the scoring or cost metric is capable of distinguising between the features of interest, but the threshold or the mechanism of utilising the score has not picked these peaks.
Using a “trial and error” approach in terms of changing parameters, we can combine a number of factors to give us the “right” answer:
# Define detector for bw=15
d1 = MovingWindow(
change_score=CUSUM(),
selection_method="detection_length",
bandwidth=15,
penalty=5
)
# Fit the detector
d1 = d1.fit(df)
# Predict / apply detector
detections = d1.predict(df)
# Preview results
detections| ilocs | |
|---|---|
| 0 | 50 |
| 1 | 150 |
The first plot shows the correct identification of the two change points and the second shows two clear peaks to indicate their location. Note that the two peaks are above the 0 in terms of the CUSUM.
Other Detectors
We’ve currently employed MovingWindow, however the skchange library also provides PELT and SeededBinarySegmentation. Both are employed with their default settings below:
PELT
# Import detector
from skchange.change_detectors import PELT# Define detector with L2Cost default
d1 = PELT()
# Fit the detector
d1 = d1.fit(df)
# Predict / apply detector
detections = d1.predict(df)
# Preview results
detections| ilocs | |
|---|---|
| 0 | 50 |
| 1 | 150 |
The first plot shows the correct identification of the two change points and the second shows a different mechanism for illustrating the score. In this instance (if you zoom in) you’ll a step change in the score itself at the two locations.
SeededBinarySegmentation
# Import detector
from skchange.change_detectors import SeededBinarySegmentation# Define detector with CUSUM default
d1 = SeededBinarySegmentation()
# Fit the detector
d1 = d1.fit(df)
# Predict / apply detector
detections = d1.predict(df)
# Preview results
detections| ilocs | |
|---|---|
| 0 | 50 |
| 1 | 150 |
The transform_scores method does not work for the SeededBinarySegmentation detector so we have to either rely on the other detectors utilising CUSUM or manually use the class to derive a suitable score that’s reflective of how it’s applied for this detector.
Conclusion
The post introduced the concept of a Change Point, which in practice may not be an individual point but rather a range of possibly correct points. Depending in the use case, they maybe sufficient of the use case of interest. Furthermore, we applied the skchange library to demonstrate how these Change Points could be detected. We demonstrated that the detection capability is strongly influenced by the choice of input parameters as well as the cost or scoring metrics.
The fundamentals for the application of Machine Learning is reinforced that the user needs to conduct a calibration activity against a known and representative time series to ensure the input parameters are appropriate for the use case to minimise erroneous results.
Version History
2025-05-29- Original2025-06-03- Minor formatting, image refresh.2025-08-31- Responsive plots and branding update.2025-09-03- Minor formatting.
Attribution
Images used in this post have been generated using multiple Machine Learning (or Artificial Intelligence) models and subsequently modified by the author.
Where ever possible these have been identified with the following symbol:

References
Citation
@online{miah2025,
author = {Miah, Ashraf},
title = {Time {Series} {Change} {Detection} {Introduction}},
date = {2025-05-29},
url = {https://blog.curiodata.pro/posts/15-time-series-change-detection/},
langid = {en}
}